|
Research Topics
|
|
Tissue specific DNA damage signaling
|
|
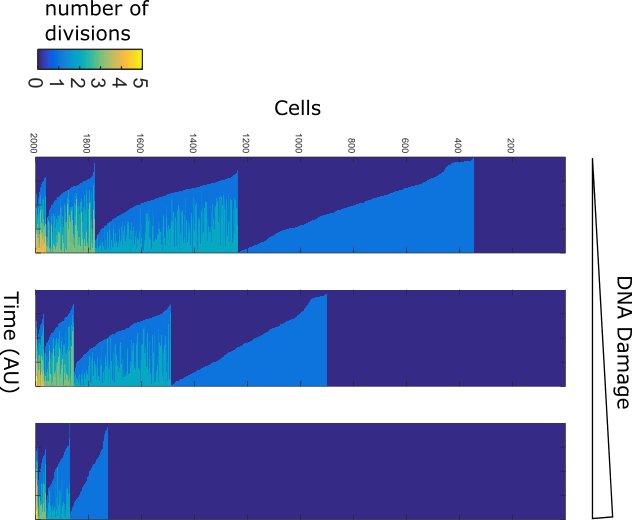
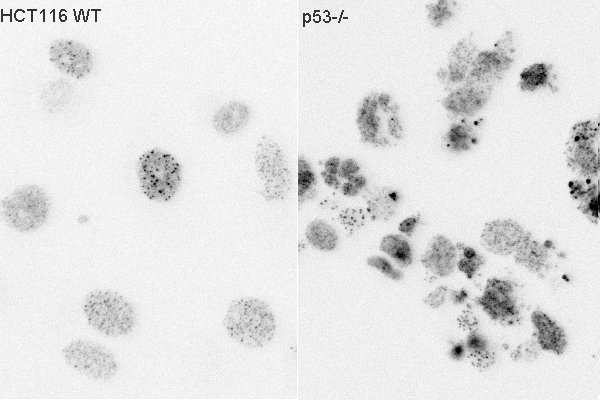
Tissues in our body show a range of radiosensitivities. Analagously some cancers are extremely senstive to chemotherapy and others are not. The DNA damage transcription factor p53 plays a role in specifying these differential sensitivities. We use in vivo (mouse) and in vitro (cell line) models of different tissues to explore how DNA damage signaling varies. We also have a long term interest in comparative DNA damage signaling between species.
|
|
Stewart-Ornstein J, Iwamoto Y, Miller MA, Prytyskach MA, Ferretti S, Holzer P, Kallen J, Furet P, Jambhekar A, Forrester WC, Weissleder R, Lahav G (2020). p53 dynamics vary between tissues and are linked with radiation sensitivity. Nat Commun. 2021 Feb 9;12(1):898.
|
|
Stewart-Ornstein J, Lahav G† (2017). p53 dynamics in response to DNA damage vary across cell lines and are shaped by efficiency of DNA repair and activity of the kinase ATM. Science Signaling 10(476).
|
|
Stewart-Ornstein J†, Cheng J, and Lahav G†(2017). Conservation and Divergence of p53 Oscillation Dynamics across Species. Cell Systems 5(4):410-417.
|
|
|
|
p53 DNA binding and regulation
|
|
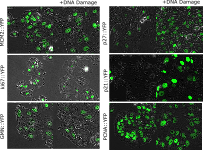
We are interested in the signaling of the DNA damage activated transcription factor p53 and specifically how it selects and regulates target genes. Though expressed in essentially all cell types p53 regulates very different gene sets in different tissue contexts. We use RNA sequencing and ChIPseq to determine p53 binding in different contexts. By applying in vitro and in vivo binding assays combined with bioinformatics and modeling we can parse the contribution of intinsic DNA sequence and chromatin state to p53 DNA binding and gene regulation.
|
|
Hafner A, Kublo L, Tsabar M, Lahav G, Stewart-Ornstein J (2020). Identification of universal and cell-type specific p53 DNA binding. BMC Mol Cell Biol. 2020 Feb 18;21(1):5.
|
|
Hafner A, Stewart-Ornstein J, Purvis J, Forester B, Lahav G† (2017). p53 pulses lead to distinct patterns of gene expression albeit similar DNA binding. Nat. Struct. Mol. Biol. 24(10):840-847.
|
|
Stewart-Ornstein J†, Nelson C, DeRisi J, Weissman JS, and El-Samad H† (2013). Msn2 coordinates a stoichiometric gene expression program. Current Biol. 23(23):2336-45.
|
|
|
|
Quiescence and DNA damage signaling
|
|
Within isogenic populations of cells the response to DNA damage and other stresses varies. We use single cell approaches such as timelapse microscopy and barcoding to study non-genetic resistance to therapy.
|
|
Stewart-Ornstein J†, Lahav G† (2016). Dynamics of CDKN1A in Single Cells Defined by an Endogenous Fluorescent Tagging Toolkit. Cell Reports, 14(7):1800-11.
|
|
Stewart-Ornstein J, Weissman JS†, El-Samad H†. (2012) Cellular Noise Regulons Underlie Fluctuations in Saccharomyces cerevisiae. Molecular Cell 45:483-93.
|
|
|